
東邦大学理学部生物学科
幹細胞リプログラミング研究室
多田�政子, PhD
リプログラミング | Masako Tada|東邦大学理学部生物学科

MASAKO TADA
Stem Cells & Reprogramming Laboratory
Our laboratory studies the mechanisms of genome-wide epigenetic reprogramming in mammalian cells, particularly in humans and mice.
DNA sequences are almost identical in all cells of each individual, but epigenetically distinct. Epigenetics gives rise to cell type-specific gene expression patterns, such as DNA methylation of cytosine in CpG sequences and modification of histone proteins responsible for regulating chromatin structure.
Epigenetics are dramatically altered during development but can be completely restored due to their reversibility. This process is called epigenetic reprogramming and is thought to be essential for acquiring nuclear pluripotency and the stable transmission of genetic information to the next generation.
We approach the process of epigenetic reprogramming by subdividing it into the following three categories:
(1) DNA cytosine methylation mechanisms in stem cells.
(2) Mechanisms of epigenetic restoration in the germline
(3) Histone exchange mechanisms in postmitotic cells (chromatin protein renewal)
Epigenetic reprogramming
Analysing phenomena in mammals using in vitro model cells.
Generation of human model cells for preclinical studies in drug discovery and development.
The following stem and embryonic cells have been generated as model cells.
(1) Transgenic mouse ESCs
(2) Transgenic human iPSCs
(3) Transgenic human hepatocellular carcinoma cell line, HepaRG and HepG2
The following stem cells and embryonic cells are used as model cells.
(1) Mouse embryonic stem cells (ESCs)
(2) Human induced pluripotent stem cells (iPSCs)
(3) Mouse epiblastic stem cells (EpiSCs)
(4) Chicken embryonic fibroblasts (CEFs)
Why is the study of epigenetic reprogramming mechanisms important?
Epigenetic reprogramming phenomena have been detected in nuclear transfer cloning, cell fusion between embryonic stem cells (ESCs) and somatic cells (our study) and induction of iPS cells from somatic cells by forced expression of four factors, Oct4, Sox2, Klf4, and cMyc. However, reprogramming is a phenomenon that always occurs twice in germ cells, even without 'induction' by special techniques, and plays an essential role in normal embryonic development. In particular, it occurs between post-fertilisation and implantation and during primordial germ cell (PGC) development. The two mechanisms are characterised by different features and are therefore referred to as embryonic and germline reprogramming, respectively. We are trying to elucidate the mechanisms of these two types of epigenetic reprogramming.
Embryonic reprogramming is essential for the nucleus to acquire pluripotency in embryonic cells and therefore they can form individuals and placenta. Germline reprogramming, on the other hand, serves to minimise genome-wide epigenetics and reset the memory of a generation. Mammalian cells contain sperm- and oocyte-derived chromosomes, and there are epigenetic differences in specific regions between the two, known as genomic imprinting. This genomic imprinting can only be erased in PGCs, minimising the differences between homologous chromosomes. This state of the PGC nucleus is called the default (initialisation, basal state) and is the most rejuvenated state of the nucleus. Thus, the significance of identifying the mechanism of epigenetic reprogramming is to understand how species maintain their genetic information across generations.
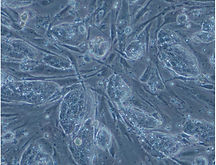
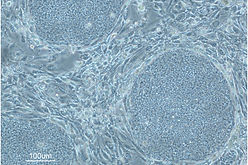
Mouse ESCs (left) and human iPSCs (right) Human iPSCs are at a more advanced developmental stage than mice.


Epigenetic reprogramming studies during embryonic development using mouse ESCs as an in vitro model and chickens as an animal model.
Embryonic reprogramming
Using the cell fusion method of somatic cells with stem cells, we have shown that embryonic reprogramming activity is always active in mouse ESCs. When somatic cell nuclei are introduced into ESCs by cell fusion, the somatic cell nuclei are reprogrammed by ESC’s factors in trans to have similar properties (epigenetics) as the ESC nuclei. Therefore, we have analysed the nature and role of the embryonic reprogramming activity of ESCs. So far, we have found that the epigenetics of the ESC nucleus itself is dynamically regulated so that it always recovers to a specific pattern. Thus, some regions are unstable, with epigenetics being erased and restored repeatedly, while others remain in an inactive state stably. Euchromatin in chromosomal arms is rich in 5-hydroxymethylcytosine, which undergoes DNA methylation and demethylation cyclically in a cell cycle-dependent manner, while pericentromeric heterochromatin is rich in 5-methylcytosine (Kubiura, M.et al., Chromosome Research, 20 (7): 837-48, 2012).
Germline reprogramming
Using a somatic cell fusion method, we found that germline stem cells EGCs derived from PGCs in female mouse E12.5 embryo have germline reprogramming activity. This function allowed the introduction of somatic cell nuclei by cell fusion to eliminate genomic imprinting of somatic nuclei.
Recently, it has become possible to generate PGC-like cells (PGC-LCs) from ESCs. Therefore, we are analysing in detail the germline reprogramming mechanisms that occur during becoming PGC-like cells from ESCs.

Epigenetic analysis using antibodies. Euchromatin localisation of 5-hydroxymethylcytosine (red) in mouse mitotic chromosomes and nuclei. Heterochromatin strongly stained blue with DAPI. Arrow; Y chromosome.
Reprogramming studies in early chicken embryos
The use of chick fertilised eggs has the advantage that epigenetic changes in early embryos can be analysed more quickly than in mice. Birds also evolved from bony fishes as tetrapods and are regarded as the survivors of extinct dinosaurs. Chickens and mammals are also included in the same group of amniotes. Therefore, comparative studies of epigenetics in chickens and mice are expected to help understand the evolutionary significance of the epigenetic reprogramming mechanisms if they can be generalised to extant bony fishes, reptiles, marsupials, and mammals. The Avian Bioscience Research Centre, affiliated with the Graduate School of Bioagricultural Sciences, Nagoya University, provided fertile eggs from the chicken and quail lineages.
In this study, a vector expressing a protein in which GFP is fused to the DNA-binding region of MBD1. This protein specifically binds to DNA methylation regions, provided by Dr Kuniya Abe of RIKEN and introduced into chicken cells to visualise DNA methylation regions in the nucleus. The figure shows live-cell imaging of mouse ESCs expressing this MBD-GFP, in which DNA methylation is accumulated in the green fluorescent areas.
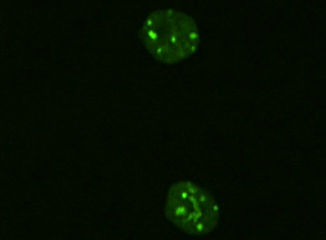
Visualisation of DNA methylation regions using MBD-GFP expression vectors.
Supporting drug discovery and development: providing replacement cells for human tissue cells
ESCs and iPSCs can differentiate into a variety of tissue cells (e.g. nerve, cardiac muscle, liver cells, pancreas). They can also proliferate continuously and are therefore expected to be a source of tissue cells for regenerative medicine. They are also expected to be a stable source of human hepatocytes that can be used for pharmacological and safety studies in drug discovery and development. Currently, primary cultures of human hepatocytes isolated from livers removed by surgery or other means are used, and this method is not authorised in Japan and currently relies on imports. In addition, thawed hepatocytes are qualitatively heterogeneous and have low proliferative potential, making it difficult to assess the safety and efficiency of the compounds. Therefore, it is expected that human hepatocyte model cells can be generated from human iPS cells to replace primary human hepatocytes. However, the current challenge is to create 'functional' hepatocytes from human iPS cells.
We considered that establishing a hepatocyte differentiation induction system would require the creation of a fluorescent reporter that would enable rapid and inexpensive quantitative assessment of the cell differentiation process in real-time. Stem cells transfected with this reporter glow red when they become hepatoblasts, the foetal hepatocytes, and green when they become adult hepatocytes. It is now possible to produce large numbers of red glowing hepatoblasts from mouse ESCs (Okuyama et al., 2020). Human iPS cells have different properties from mouse ESCs, and therefore, the differentiation methods of hepatocytes must also differ. Human iPS cells are at a different developmental stage from mouse ESCs and should be treated as early epiblast cells.

